Transcriptomics vs. Lipidomics
The purpose of analyzing transcriptome and lipidome data.
The biological process regulation trajectory to the gene transcription level is often consider the figure of biological phenotype.
In that scenario, mRNA expression pattern is recognized as the “biomarker” of biological process.However, in the complex regulation
of lipid metabolism, the causal relation of gene regulation and abundant of lipid metabolites are often difficult to be illustrated.
The major setback bottleneck for researchers are the varied format between Next Generation Sequencing (NGS)-based transcriptome
and Liquid Chromatography-Mass Spectrometry (LC-MS) based lipidome data. TBAL have fruitful experience helping biomedical
scientist in integrating various format of data,and make sense of them.
Here’re some example:
Legend:
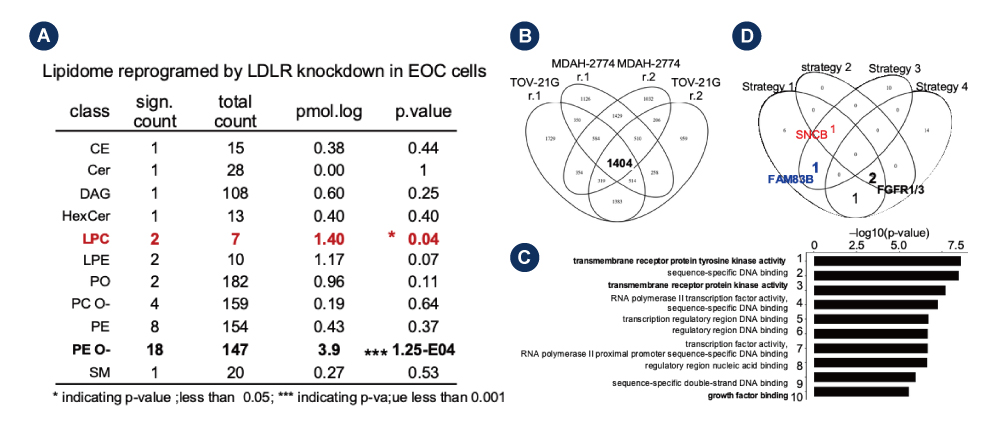
Transcriptome vs. lipidome trans-omics approaches for potential molecular regulations.
A.Lipidome analyses of lipid species comparing MDAH-2774 and TOV-21G cells with or wo/ treatments.
B. Replicated transcriptome analysis by RNAseq was performed to compare MDAH-2774 and TOV-21G cells with or wo/ treatments.
C.Pathway enrichment analysis by GO-term followed with GSEA analysis. The top-10 enriched pathways were ranked from highest to lowest p-value(-log10).
D.Trans-omics analyses of transcripto2me and lipidome profiles. The four selection criteria were implemented in the analyses.
(Endocrine-Related Cancer,Chang et al, 2020; doi.org/10.1530/ERC-19-0095)
Reference:
1. Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R. and Salzberg, S.L. (2013) TopHat2: accuratealignment of transcriptomes in the presence of insertions,
deletions and gene fusions. Genome Biol, 14,R36.
2. Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D.R.,Pimentel, H., Salzberg, S.L., Rinn,J.L. and Pachter, L. (2012) Differential gene and transcript
expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols, 7,562-578.
3. Chung, I.F., Chen, C.Y., Su,S.C., Li, C.Y., Wu, K.J., Wang, H.W. and Cheng, W.C. (2016) DriverDBv2: a database for human cancer driver gene research.
Nucleic Acids Res, 44,D975-979.
4. Cheng, W.C., Chung, I.F., Chen, C.Y., Sun,H.J., Fen, J.J., Tang, W.C., Chang, T.Y., Wong, T.T. and Wang, H.W. (2014) DriverDB: an exome sequencing database
for cancer driver gene identification. Nucleic Acids Res, 42,D1048-1054.
5. Kanehisa, M., Goto,S., Sato, Y., Furumichi, M. and Tanabe, M. (2012) KEGG for integration and interpretation of large-scale molecular data sets.
Nucleic Acids Res, 40,D109-114.
6. Schaefer, C.F., Anthony, K., Krupa, S., Buchoff, J., Day, M., Hannay, T. and Buetow, K.H. (2009) PID: the Pathway Interaction Database.
Nucleic Acids Res, 37,D674-679.
7. Croft, D., O'Kelly, G., Wu, G., Haw, R.,Gillespie, M., Matthews, L., Caudy, M., Garapati, P., Gopinath, G., Jassal, B. et al. (2011) Reactome: a database of reactions,
pathways and biological processes. Nucleic Acids Res, 39,D691-697.
8. Subramanian, A., Tamayo, P., Mootha, V.K., Mukherjee, S., Ebert, B.L.,Gillette, M.A., Paulovich, A., Pomeroy, S.L., Golub, T.R.,Lander, E.S.et al. (2005)
Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A, 102,15545-15550.
想跟我們一對一交流嗎?
歡迎留下您的問題,我們將進一步聯繫您。